Biology and Biotechnology Program, L-452
Lawrence Livermore National Laboratory
Livermore, California 94551
Abstract
A number of neurodegenerative diseases have been found to be
strongly associated with proteins containing a polyglutamine
stretch which is greatly expanded from approximately 20
glutamines in normal individuals to over 40 in affected
individuals. A conformational change in the expanded
polyglutamine stretch is believed to form the molecular basis for
disease onset. Model peptides containing glutamine repeats tend
to aggregate and to become insoluble. We have synthesized readily
water soluble monomeric peptides by flanking polyglutamine
stretches with sequences rich in alanine and lysine. Circular
dichroism measurements show that polyglutamine stretches of
length 9 or 17 adopt a random coil configuration in aqueous
solution. Our method to design soluble monomeric peptides
containing extended polyglutamine stretches may be generally
useful in studying other highly aggregating peptides.
Introduction
Over 30 years ago Krull and colleagues (1) provided the first structural information on polyglutamine. Using a number of experimental techniques and molecular modeling, they showed that peptides made of polyglutamine will form large aggregates with the individual peptides of the aggregates joined in b-sheets with the side chain amides of the glutamines participating in hydrogen bonds with other side chain amides and/or main chain amides. Since then a number of other groups have come to similar conclusions about the structure of polyglutamine containing peptide aggregates (2,3). Krull and colleagues were interested in polyglutamine as a model system to study proteins such as wheat gluetin which has a high percentage of glutamine.
Recently, polyglutamine has re-emerged as a model, - this time, however, in a tragic context: five neurodegenerative diseases (Huntington disease (HD), spinocerebellar ataxia type 1 (SCA1), dentatorubral-pallidoluysian atrophy (DRPLA), Macado-Joseph disease (MJD), and spinobulbar muscular atrophy (SBMA)) each have been found to be strongly associated with a protein containing a polyglutamine stretch which is greatly expanded in affected individuals (see ref. 4 for a review). For the five diseases, the mean length of the glutamine repeat in unaffected individuals is approximately twenty, and the cutoff for pathology is about forty (the cutoff may be higher for MJD (4)). Furthermore, long polyglutamine stretches have been found in many transcription factors (5).
Based on the ability of polyglutamine containing proteins and peptides to aggregate (1-3, 6), it has been suggested that the pathology in HD and the other expanded polyglutamine stretch associated diseases is caused by some kind of anomalous binding activity of an expanded polyglutamine stretch in one protein to a polyglutamine stretch or other region in another protein. However, in studies of the proteins associated with HD by a number of groups (7,8), the protein associated with SCA1 (9), and the protein associated with DRPLA (10) , no dimerization of polyglutamine containing proteins, or aberrant binding of a protein containing an expanded polyglutamine stretch to another protein has yet been observed.
Results and Discussion
Given the similar cutoff number of glutamines for pathology in the different diseases and the current lack of observations of aberrant binding activity of expanded polyglutamine stretches, we became interested in the structure of polyglutamine in a monomeric peptide. Specifically, we speculated whether longer stretches of poly-Q might adopt different conformations than shorter ones. It has thus far has been difficult to study monomeric peptides containing long polyglutamine stretches because of their tendency towards aggregation and insolubility. For example, the peptide Ac-D2Q15K2-NH2 rapidly becomes insoluble in aqueous solution at neutral pH and is thought to form b-sheet aggregates (3). It is conceivable that the acidic and basic ends of this peptide form intermolecular salt bridges and thus promote association or b - sheet formation. To exclude this possibility, we synthesized a similar peptide, Ac-K2Q17YK2-NH2, with basic residues on both ends. This peptide was readily soluble in aqueous solution at neutral pH, but ultracentrifugation analysis showed that it formed large aggregates (data not shown) and the CD spectrum of this peptide strongly resembled the spectrum of the peptide Ac-D2Q15K2-NH2 .
Baldwin and colleagues constructed a basic peptide
Ac-(A4K)3A-NH2 which was 80% a-helical in aqueous solution (11).
We reasoned that by flanking a long polyglutamine stretch with
sequences using the -(A4K)- motif of Baldwin and colleagues we
might be able to obtain a soluble, monomeric peptide containing
an extended polyglutamine stretch. We synthesized two peptides,
AKQ9 (Ac-(A4K)3Q9KA4KA-NH2) and AKQ17
(Ac-YGA2KA4KQ17KA4KA-NH2), containing 9 and 17 glutamines
respectively. These two peptides are readily soluble in aqueous
solution at neutral pH, and results from ultracentrifugation show
that the peptides are in fact monomeric. Using circular dichroism
(CD) spectroscopy we have found that the polyglutamine stretches
in these
peptides adopt a random coil configuration.
The CD spectra (Fig.1) for AKQ9 and AKQ17 with increasing amounts of trifluoroethanol (TFE), a known a-helix inducer, are similar in their qualitative appearance and both show well defined isometric points indicative of a two state system. The spectra were analyzed using a linear combination fit based on model data (12).
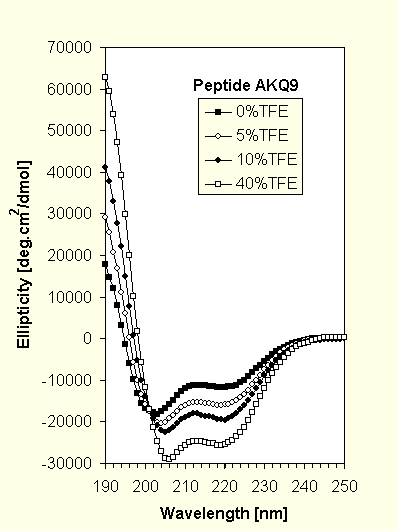
AKQ9 in aqueous solution consists of 41% helix and 59% random coil. We assume that not all of the flanking Baldwin peptide is helical, most likely at the termini and in the transition region to the poly-Q stretch. Adding TFE increases the helix content (the effect saturates at about 40% TFE) and the amount of coil remaining can be nearly quantitatively attributed to the 9-residue poly-Q stretch (71% helix, 29% coil).
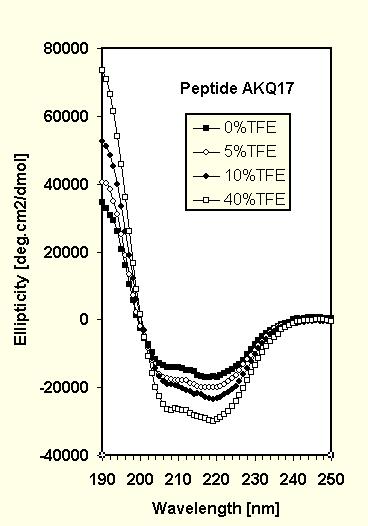
For AKQ17 in aqueous solution, the fit yields a larger amount of random coil (67%) than for AKQ9, which is in agreement with the longer 17-residue poly-Q stretch. The remainder consists of 26% helix and a small amount of b-sheet (7%). Such an amount of b-sheet represents only about 3 residues and is of the order of uncertainty for a CD spectrum fit, although it may indicate some nascent hairpin forming ability. AKQ17 responds to the addition of TFE at the expense of all sheet and some random coil (64% of helix, 36% coil at 40% TFE), with most of the poly-Q remaining in a coil conformation.
For a number of reasons we infer that the alanine/lysine rich regions are helical while the polyglutamine stretches adopt random coil configurations. First, the N-terminal 15 amino acids of AKQ9 are the first 15 out of 16 amino acids of the peptide of Baldwin and colleagues, which was shown to be strongly helical in aqueous solution, and the other N-terminal of AKQ9 and the C-terminal regions of AKQ9 and AKQ17 use the -(A4K)- motif of Baldwin. Second, the percentage of helix decrease and of coil increase from AKQ9 to AKQ17 parallels the decrease of alanine/lysine content and increase in glutamine content. Finally, working with poly-[N5-(hydroxyalkyl-glutamine)] compounds, Lupu-Lotan and colleagues (14) found that poly-[N5-(hydroxybutyl-glutamine)] was strongly helical in aqueous solution, poly-[N5-(hydroxypropyl-glutamine)] was mixed helix and coil, and poly-[N5-(hydroxyethyl-glutamine)] was all coil. A coil conformation for poly-[N5-(hydroxyethyl-glutamine)] was confirmed by Alder et al. (13). Following this trend of increasing coil content with shorter alkyl chains, one would expect polyglutamine itself to be a coil rather than a helix.
Summary
We have shown that the polyglutamine stretches in the water
soluble peptides AKQ9 and AKQ17 adopt a random coil conformation.
In the peptide Ac-D2Q15K2-NH2, and in proteins in which eleven
consecutive glutamines have been inserted into the inhibitory
loop of a truncated chymotrypsin inhibitor 2, it has been found
that the polyglutamine stretch forms a hairpin (3,6). These
results taken together with ours demonstrate the ability of
polyglutamine to adopt different conformations when flanked by
different sequences. Since the cutoff number of about 40
consecutive glutamines for disease pathogenesis seems to be
common across the diseases, it is tantalizing to speculate that
above some number of glutamines polyglutamine no longer adopts a
random coil conformation, but forms a hairpin (15). Potential
drugs could stabilize an extended chain conformation or block
hairpin formation. Alternatively, it might be possible to induce
cells to incorporate N5-(hydroxyalkyl-glutamine) into long
glutamine repeats to stabilize a coil conformation.
References
1. L.H.Krull et al., Biochemistry 4, 626 (1965).
2. K.Ishikawa and T.Endo, Journal of Polymer Science: Part A:
Polymer Chemistry, 28, 3525-3527 (1990) .
3. M.F.Perutz et al., Proc. Natl. Acad. Sci. U.S.A. 91, 5355-5358
(1994).
4. D.E.Housman, Nature Genet. 8, 10-11 (1995).
5. H.P.Gerber et al., Science 263, 808-811 (1994).
6. K.Stott et al., Proc. Natl. Acad. Sci. U.S.A. 92, 6509-6513
(1995).
7. Y.S.Jou and R.M.Myers, Human Molec. Genet. 4, 465-469 (1995).
8. Y.Trottier et al., Nature Genet. 10, 104-110 (1995).
9. A.Servadio et al., Nature Genet. 10, 94-98 (1995).
10. Y.Yazawa et al., Nature Genet. 10, 99-103 (1995).
11. S.Marqusee, V.H.Robbins, R.L.Baldwin, Proc. Natl. Acad. Sci.
USA 86, 5286-5290 (1989).
12. N.Greenfield and G.D.Fasman, Biochemistry 8, 4108-4116
(1969).
13. A.J.Adler et al., J. Amer. Chem. Soc. 90, 4736-4738 (1968).
14. N.Lupu-Lotan, A.Yaron, A.Berger, Biopolymers 4, 365-368
(1966).
15. See also C. Jennings, Nature 378, 127 (1995), and Y. Trottier
et al., Nature 387, 403-405 (1995).
16. We thank G.D. Fasman, D. Major, and M. Brody for helpful
discussions, M.Maestre for invaluable technical assistance, and
Y.-R. Yang for ultracentrifugation analysis. Work performed at
Lawrence Livermore National Laboratory under the auspices of the
U.S. DOE under Contract W-7405-ENG-48.
Notes
The peptides were prepared in-house using a PS3 automated
solid phase peptide synthesizer (Protein Technologies Inc.,
Woburn, Ma). On completion of the synthesis the free N terminus
was acetylated with acetic anhydride. The peptide was cleaved
from the Rink amide linker with trifluoroacetic
acid/thioanasole/triethylsilane and purified to homogeneity by
preparative HPLC (PRP-2 column (Hamilton Co., Reno, NV)).
Presence of the peptides was confirmed by electrospray mass
spectroscopy: expected for AKQ9 3061.5, found 3061.08 (0.98);
expected for AKQ17 3751.9, found 3751.87 (0.14) g/mol. Molecular
weights of the peptides dissolved in 50mM phosphate, pH 7.0, were
determined using a Beckman Model XL-A analytical ultracentrifuge
operating in sedimentation equilibrium mode at 50000 rpm and at
20oC. The absorbance was recorded at 276 and 235 nm, and the data
fitted with a Levenberg-Marquart minimization to an ideal single
component model using a solvent density of 1.0 g/cm3 and a
partial specific particle volume (v) of 0.707 cm3/g; found
molecular weights for AKQ9, 3140 (82) g/mol; and for AKQ17, 3719
(74) g/mol. CD spectra were recorded at room temperature using a
computerized Cary model 60 spectropolarimeter (5mM phosphate
buffer , pH 7.0, 0.5 mm cells). Circular Dichroism is reported as
mean residue ellipticity in degee.cm2/dmol. The spectra were
analyzed by a linear combination fit using the reference data of
Greenfield and Fasman (12).
Back to CD tutorial
![]() Back to X-ray Facility Introduction
Back to X-ray Facility Introduction
LLNL Disclaimer
This World Wide Web site conceived and maintained by
Bernhard Rupp (br@llnl.gov)
Last revised April 06, 1999 11:02
UCRL-MI-125269