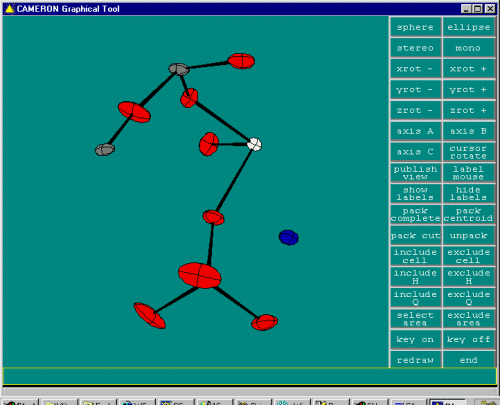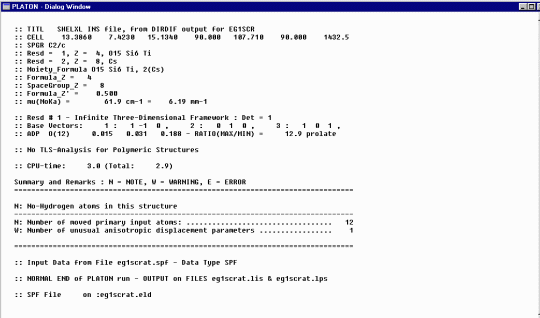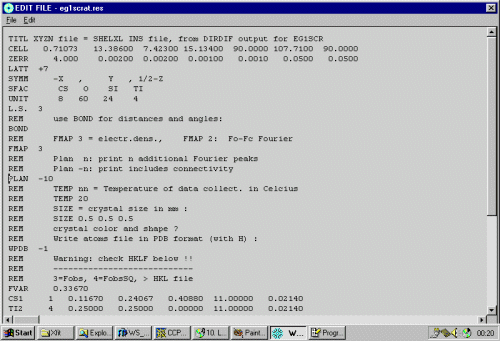
To do some cycles of structure refinement, select Refine, SHELXL-97, in this case, using the Analytically Absorption Corrected data.
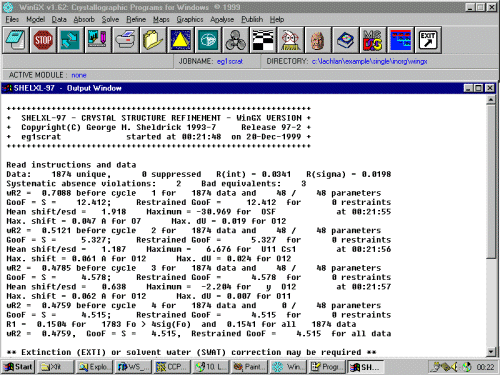
(Note: This run-through glosses over graphically looking at the structure and evaluating the bond-length and angles list which would normally be occuring.) Looking at the results, the thermal on O14 is very large, implying it could be a bogus atom. Delete this and extend the number of Least Squares cycles to 9 by changing L.S. 4 to L.S. 9
CS1 1 0.117374 0.241669 0.409920 11.00000 0.03160 TI1 4 0.250000 0.250000 0.000000 10.50000 0.01444 SI3 3 -0.002245 0.231140 -0.146855 11.00000 0.01286 SI4 3 0.324271 0.429524 -0.166738 11.00000 0.01928 SI5 3 0.345265 0.040898 -0.144329 11.00000 0.02120 O6 2 0.381402 0.240320 -0.169454 11.00000 0.03022 O7 2 0.301623 0.051163 -0.057210 11.00000 0.02355 O8 2 0.107629 0.189508 -0.074593 11.00000 0.02792 O9 2 0.253355 -0.037705 -0.230301 11.00000 0.04742 O10 2 0.000000 0.250040 -0.250000 10.50000 0.07091 O11 2 0.236178 0.075979 0.090143 11.00000 0.02842 O12 2 0.418745 0.564177 -0.151408 11.00000 0.06600 O13 2 0.451799 -0.079446 -0.128166 11.00000 0.04612 O14 2 0.367496 0.229404 -0.245969 11.00000 0.19651