TI1 3 0.000000 0.000000 0.000000 10.25000 0.05811
TI2 3 1.144282 0.500000 0.520498 10.50000 0.03427
O3 2 1.094371 0.000000 0.256022 10.50000 0.05397
O4 2 1.227591 1.000000 0.687662 10.50000 0.13380
O5 2 1.081518 0.500000 0.899902 10.50000 0.00001
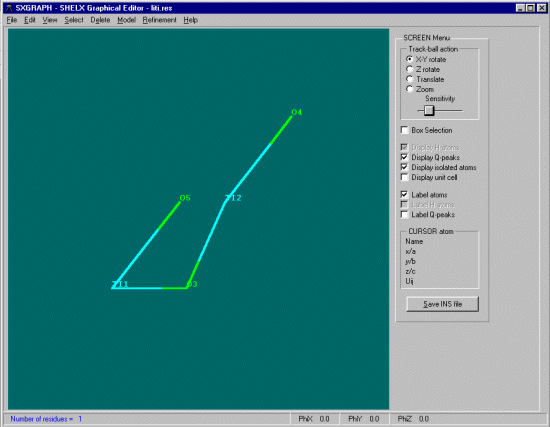
Opening up the Shelx INS file in cameron shows how the atoms in this structure are in separate parts of the cell, but it would be nice if the unique atoms could be is such positions as they bond nicely for convenient viewing and such exercises as slant plane fourier maps defined by the position of 3 unique atoms.
To enter the SXGRAPH (Primarily a GUI interface for handling Shelx *.RES and *.INS files), select: Refine, SXGRAPH.
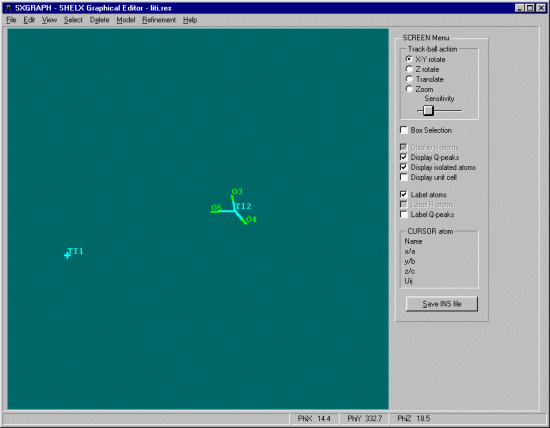
Within SXGRAPH, select Model, Assemble Residues after which WinGX will do it's duty to this structure.