Crystallography of microsomal membrane-bound P450 which we have
recently solved (January issue of Molecular Cell) and have launched a major
new program to study the structure and function of. P450s are involved
in metabolism of numerous drugs, xenobiotics and carcinogens. Until now
all work has been based on homologous bacterial structures. With the solution
of microsomal P450 we are in a position to make major new advances in this
field.
48. Williams, P.A., Cosme, J., Sridhar, V., Johnson, E.F., McRee, D.E.
(2000) The Crystallographic Structure of a Mammalian Microsomal Cytochrome
P450 Monooxygenase: Structural adaptations for Membrane Binding and Functional
Diversity. Molecular Cell, in press (January).
High-resolution studies of metalloprotein centers. In this project
we are doing the structures of metal centers at high to very high resolutions
in order to determine the exact geometry of various metal centers upon
functional change. Central to this project is the use of SHELX to determine
the standard uncertainties. Since metals diffract strongly, we can determine
the metal-ligand bonds to one-hundredth of an Ångstrom and thus are
in a position to make strong statements about metal geometry and changes.
References:
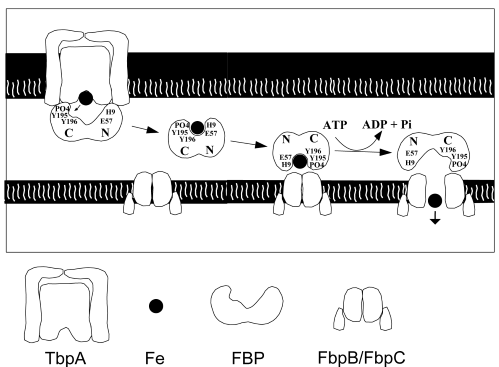
Uptake of iron in pathogenic bacteria. This porject has
is elucidating the pathway of iron uptake into several calasses of pathogenic
bacteria with the long term goal of producing new antibacterial agents
and for understanding uptake through membranes in general. We have
solved the structure of the major component of the system ferric binding
protein (FBP) and are now in the process of elucidating the function of
FBP in binding iron and its interactions with the mebrane bound components
(see diagram below):

Reference. Bruns, C.M., Nowalk, A.J., Arvai, A.S., McTigue, M.A., Vaughan, K.G., Mietzner, T.A. and McRee, D.E. (1997) Structure of H. influenzae Fe+3-binding protein: Convergent evolution within a superfamily. Nature Struct. Biol. 4, 919-924.
High-throughput structure determination. As part of a major structural genomics effort we are working on methods to automate and improve high-throughput structure determinations. Some programming skills are needed for this job. TSRI offers increased compensation to programmers in recognition of the high demand for their skills.
TSRI offers an excellent environment for structural biology with the latest in equipment and computers. The La Jolla/San Diego area is an excellent place in which to live, with some of the finest weather and scenery in the world.
Please send CV and references to: dem@scripps.edu