The Example described here is 3yr4 from the supplied examples (which must be installed first).
Start Oscail click the ![]() button,
set Files of type to All files and navagate to the 3yr4
example which should be in \Xlearn.
button,
set Files of type to All files and navagate to the 3yr4
example which should be in \Xlearn.
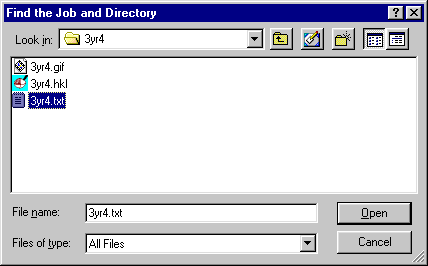
select any 3yr4 file and click Open.
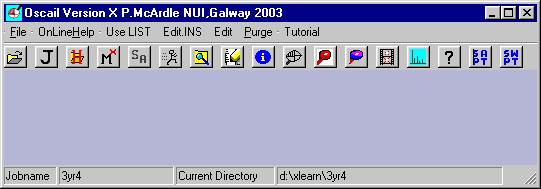
The correct Jobname and Current Directory should be on the status bar.
The first task is to determine Z (the number of molecules in the unit cell, the crystal system and the space group. Select Edit / Edit.TXT.
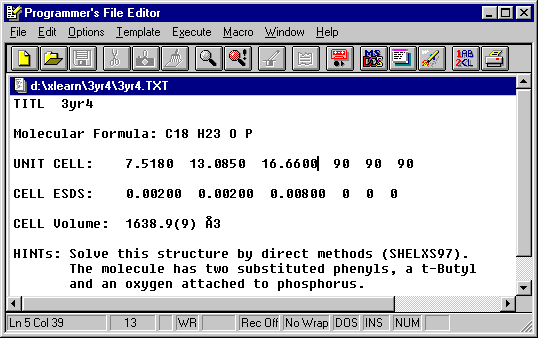
Z is estimated using the approximation that each non-H
atom occupies 20Å3.
Thus Z = 1638.9 / (20 x 20) = 4 and the crystal system is orthoroombic.
Close the PFE editor window. Absen is used the decide the space group. Click ![]() and select Absen and OK
and select Absen and OK
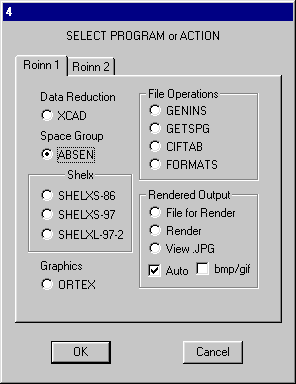
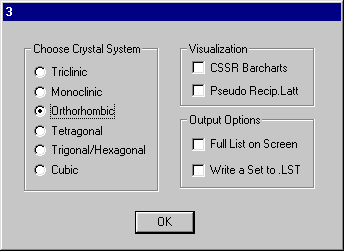
Select Orthorhombic and OK.
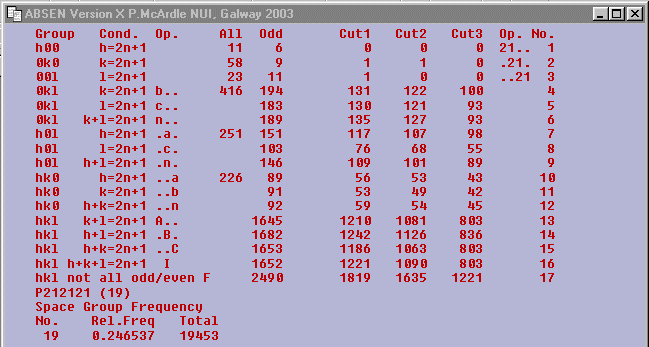
The space group is uniquely determined to be P212121
No. 19.
There is now sufficient information to solve the structure using direct methods.
Run Genins as you ran Absen (it is on the top right of the
dialog).
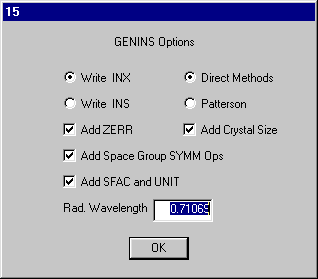
The defaults are all suitable click OK and OK on the comment entry dialog (2).

Enter the cell dimensions and click OK
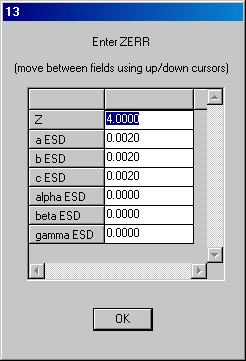
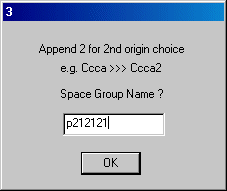
Z is correct so click OK.Click OK to the next dialog and enter the space group name on dialog 3 and OK.
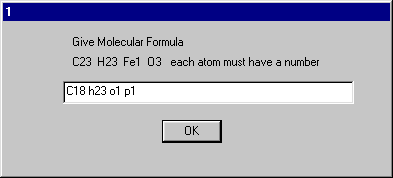
The molecular formula must be entered correctly.
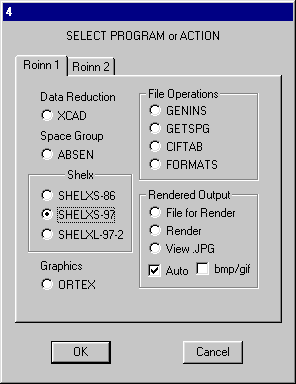
Click OK and the mean volume per non-H atom is computed this should be reasonable and it is a check on the entered data. Click OK on dialog 13 and OK to exit. As suggested in the hints you must use SHELXS-97 select this and OK.
Select Yes on Review Screen Output.
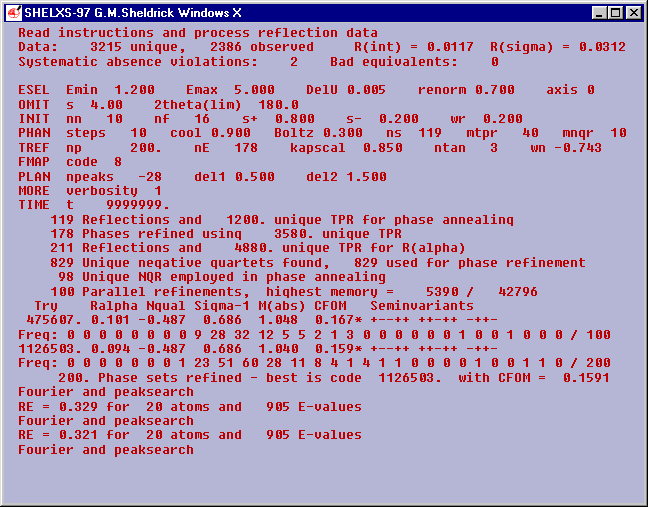
The best CFOM is 0.1591 (a bit high but it works).
Click the screen again and Select NO on Review Screen Output. To examine the
structure click ![]() to
run Ortex. Select Yes on Use Defaults. In Ortex Setup select NO Atoms in Line Mode and click to show atom labels
to
run Ortex. Select Yes on Use Defaults. In Ortex Setup select NO Atoms in Line Mode and click to show atom labels ![]() and the following picture
will appear.
and the following picture
will appear.
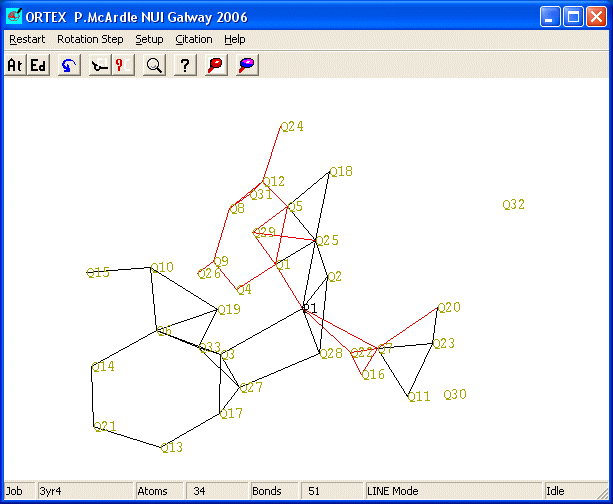
The task now is to delete rubbish and rename any non-C
atoms. The principle is to DELETE update and RENAME in that order. By default
unidentified atoms are labelled Q and treated C
atoms. Rubbish atoms are in unreasonable positions and often have high numbers i.e.
> the number of non-H atoms. Common chemical sense (three membered rings are
rare for example) can sort out most of the problems. After reading the hints
rubbish was deleted to leave the following. To delete atoms enter edit mode
by clicking ![]() and in edit
a selected atom can be deleted using the D key on the keyboard. The
currently selected atom is indicated on the status bar. To select and atom left click the centre left of the atom name.
and in edit
a selected atom can be deleted using the D key on the keyboard. The
currently selected atom is indicated on the status bar. To select and atom left click the centre left of the atom name.
There are 20 non-H atoms in the structure. Delete all atoms which have numbers greater than Q21 and you should have.
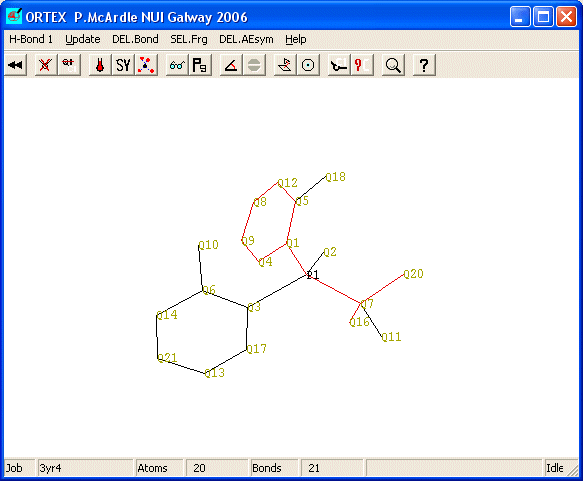
It is necessary to save the changes to do this click the
right mouse button and to Add Changes select Yes and Yes on
Use Defaults. The changes are now saved but it is necessary to go into
Edit again to rename the Oxygen on the Phosphorus. Select the oxygen and press C
to rename enter the name O1 (case does not matter) and click OK.
Click the right mouse button to save the changes.
To switch all the Qs to Cs and set unique atom numbers click Restart
on the Ortex menu and NO to Use Defaults, Yes to Use
Covalent Radii.
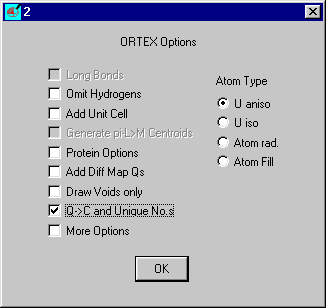
and on the Ortex Options Select Q -> C and Unique No.s and OK. Exit Ortex and run SHELXL-97/2.
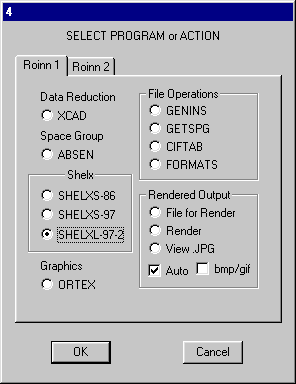
Click OK and the structure will be refined. R1 will
now be at 0.2564. Select Yes on Overwrite INS dialog and NO
to review Screen Output. Now run Ortex again, select the defaults
and the structure will appear.
Click ![]() to enter atom
mode.
to enter atom
mode.
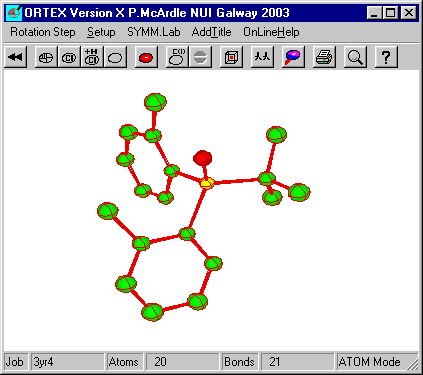
Click the Printer icon on the toolbar and select PNG (1200
x 900) and OK
If you insert this PNG file into WORD you should get
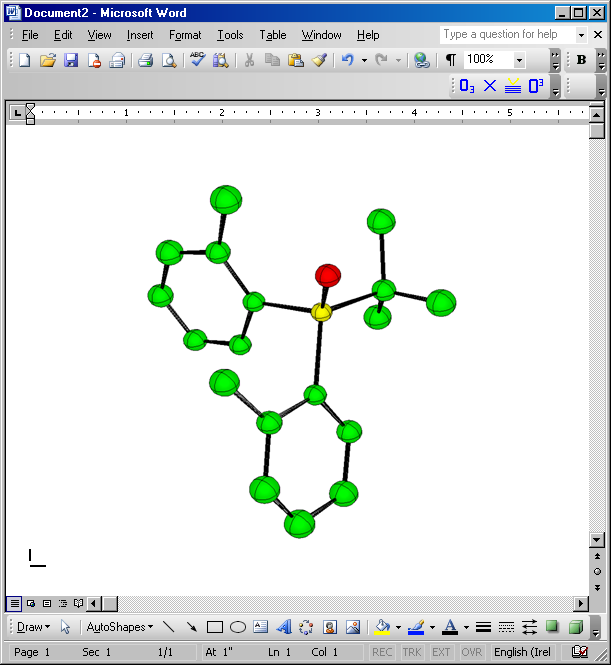
More work (using anisotropic parameters and putting in H atoms) will reduce the R factors further. Many different types of pictures can be easily produced.