



Next: The AUTO wrapper.
Up: Using the program
Previous: Supported crystallographic calculations
Contents
The .mtz wrapper
GraphEnt offers limited capabilities for a completely automated run with only input the name
of a .mtz file. The major limitation is that you do not assign columns or chose a type of
calculation. What happens is that GraphEnt will open your .mtz file and read the list of
column types. If it finds a recognisable set of column types it will simply go ahead and
do the calculation. If what GraphEnt decides to do is not what you wanted, simply use
mtzutils or sftools to select, or create the column types that GraphEnt expects for
your type of calculation.
This is the list of column types ( order is important)
and corresponding calculation that
GraphEnt will perform :
| Column types |
Action performed |
| HHHFQPW |
Assumed to be
h, k, l, F,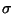 (F), (F),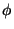 , FOM. Synthesis will be
mFexp(i , FOM. Synthesis will be
mFexp(i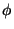 ) ) |
| HHHFQP |
Assumed to be
h, k, l, F,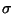 (F), (F),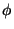 . Synthesis will be
Fexp(i . Synthesis will be
Fexp(i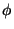 ) ) |
| HHHFQFQDQ |
Assumed to be
h, k, l, FP,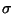 (FP), FPH, (FP), FPH,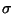 (FPH), (FPH),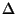 Fano, Fano,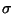 ( (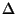 Fano) for a derivative. Synthesis will be
a
(FP - FPH)2 isomorphous difference Patterson function.
An input file for the anomalous synthesis will also be
prepared (which can be used as input for a second run). Fano) for a derivative. Synthesis will be
a
(FP - FPH)2 isomorphous difference Patterson function.
An input file for the anomalous synthesis will also be
prepared (which can be used as input for a second run). |
| HHHFQFQ |
Same as the previous one, but no additional input
file for the anomalous part is prepared. |
| HHHDQ |
Assumed to be
h, k, l,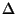 Fano, Fano,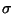 ( (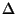 Fano). Synthesis will be an anomalous difference Patterson function
( Fano). Synthesis will be an anomalous difference Patterson function
(
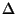 Fano2). Fano2). |
| HHHFQ |
Assumed to be
h, k, l, F,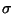 (F). Synthesis will be the
Patterson function. (F). Synthesis will be the
Patterson function. |
Please note that GraphEnt will only check the column types immediately after the indeces, and if a
match is found the rest of the columns will be ignored. Furthermore, the search is performed
in the order shown in the table and the calculation performed is the first matching. What
this means is that if your column types are HHHFQP, GraphEnt will go for a phased synthesis
no matter what you may wanted to do. If your intention was to calculate a Patterson function
with Fs and 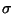 (F)s, you will have to use mtzutils or
sftools to remove the column containing the phases.
(F)s, you will have to use mtzutils or
sftools to remove the column containing the phases.
If the column types of your .mtz file are in the right order, just give
GraphEnt <my_file.mtz> for a run that will use all data present in the file, or
GraphEnt 15.0 3.5 <my_file.mtz> to use only data between 15 and 3.5Å resolution.
If what you are calculating is isomorphous difference Patterson functions for a derivative,
and your space group has centric zones of the type
h0l, hk0, 0kl, you may as well try
something like GraphEnt h0l 15.0 3.5 <my_file.mtz> to calculate the [010] Patterson
projection. GraphEnt will also recognise and use all zone selections recognised
by mtzutils (ie H00, 0K0, 00L, HH0, -HH0, HHH, HK0, 0KL, H0L and HHL).
NOTE WELL : GraphEnt is always performing the calculation in space group P1.
To avoid unnecessary repetition, the program calls CAD (and possibly
MTZUTILS) from the CCP4 distribution to expand the data to P1. This
means that GraphEnt will fail if CCP4 is not correctly set-up or if the various
symbols are not defined (especially the CLIBD variable, you can check
its presence with setenv | grep 'CLIBD').
NOTE WELL : Maximum entropy maps may well predict
non-zero amplitudes for data beyond the high resolution
limit of your input data set (thus giving --for good data-- a degree of ``super-resolution''10).
For this reason the grid size that GraphEnt uses is significantly
larger than that used by the conventional FFT (and even this may not be large enough).
The implication is that if you want to do a calculation using all your data
to 2Å resolution, you are better-off submitting a batch job for the night
instead of trying to do it interactively.
In addition, and because
GraphEnt is doing the calculation in P1, the higher the symmetry, the larger the grid, the
slower GraphEnt will be. As a last precaution, I should add that I have never performed a
calculation with more than 9437184 (=192x256x192) pixels.
Footnotes
- ... ``super-resolution''10
-
Hopefully, and if the F000 and standard deviations of your data are correctly estimated, the maxent
map should only show you the degree of resolution that is required by your data. The fact that it may
look sharper than the conventional map is not due to a ``super-resolution'' effect arising from the
maxent algorithm, but because the conventional transform is not optimal for your problem. To make this
clear, if you had a 100% complete and noise-free data set extending to infinite resolution,
then the conventional and maxent map would be
identical. Actually, if you start using maxent systematically, you will note that the conventional and
maxent maps start looking quite similar under considerably less stringent conditions.




Next: The AUTO wrapper.
Up: Using the program
Previous: Supported crystallographic calculations
Contents
NMG, Nov 2002